# Import the libraries we need for this lab
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
import torch
from torch.utils.data import Dataset, DataLoader
import torch.nn as nnLogistic Regression and Bad Initialization Value
Training Two Parameter, Mini-Batch Gradient Decent, Training Two Parameter Mini-Batch Gradient Decent

Objective
- How bad initialization value can affects the accuracy of model. .
Table of Contents
In this lab, you will see what happens when you use the root mean square error cost or total loss function and select a bad initialization value for the parameter values.
- Make Some Data
- Create the Model and Cost Function the PyTorch way
- Train the Model:Batch Gradient Descent
Estimated Time Needed: 30 min
Preparation
We’ll need the following libraries:
Helper functions
The class plot_error_surfaces is just to help you visualize the data space and the Parameter space during training and has nothing to do with Pytorch.
# Create class for plotting and the function for plotting
class plot_error_surfaces(object):
# Construstor
def __init__(self, w_range, b_range, X, Y, n_samples = 30, go = True):
W = np.linspace(-w_range, w_range, n_samples)
B = np.linspace(-b_range, b_range, n_samples)
w, b = np.meshgrid(W, B)
Z = np.zeros((30, 30))
count1 = 0
self.y = Y.numpy()
self.x = X.numpy()
for w1, b1 in zip(w, b):
count2 = 0
for w2, b2 in zip(w1, b1):
Z[count1, count2] = np.mean((self.y - (1 / (1 + np.exp(-1*w2 * self.x - b2)))) ** 2)
count2 += 1
count1 += 1
self.Z = Z
self.w = w
self.b = b
self.W = []
self.B = []
self.LOSS = []
self.n = 0
if go == True:
plt.figure()
plt.figure(figsize=(7.5, 5))
plt.axes(projection='3d').plot_surface(self.w, self.b, self.Z, rstride=1, cstride=1, cmap='viridis', edgecolor='none')
plt.title('Loss Surface')
plt.xlabel('w')
plt.ylabel('b')
plt.show()
plt.figure()
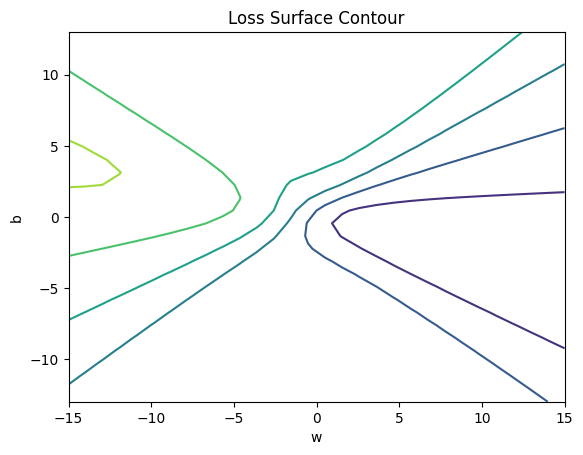
plt.title('Loss Surface Contour')
plt.xlabel('w')
plt.ylabel('b')
plt.contour(self.w, self.b, self.Z)
plt.show()
# Setter
def set_para_loss(self, model, loss):
self.n = self.n + 1
self.W.append(list(model.parameters())[0].item())
self.B.append(list(model.parameters())[1].item())
self.LOSS.append(loss)
# Plot diagram
def final_plot(self):
ax = plt.axes(projection='3d')
ax.plot_wireframe(self.w, self.b, self.Z)
ax.scatter(self.W, self.B, self.LOSS, c='r', marker='x', s=200, alpha=1)
plt.figure()
plt.contour(self.w, self.b, self.Z)
plt.scatter(self.W, self.B, c='r', marker='x')
plt.xlabel('w')
plt.ylabel('b')
plt.show()
# Plot diagram
def plot_ps(self):
plt.subplot(121)
plt.ylim
plt.plot(self.x, self.y, 'ro', label="training points")
plt.plot(self.x, self.W[-1] * self.x + self.B[-1], label="estimated line")
plt.plot(self.x, 1 / (1 + np.exp(-1 * (self.W[-1] * self.x + self.B[-1]))), label='sigmoid')
plt.xlabel('x')
plt.ylabel('y')
plt.ylim((-0.1, 2))
plt.title('Data Space Iteration: ' + str(self.n))
plt.show()
plt.subplot(122)
plt.contour(self.w, self.b, self.Z)
plt.scatter(self.W, self.B, c='r', marker='x')
plt.title('Loss Surface Contour Iteration' + str(self.n))
plt.xlabel('w')
plt.ylabel('b')
# Plot the diagram
def PlotStuff(X, Y, model, epoch, leg=True):
plt.plot(X.numpy(), model(X).detach().numpy(), label=('epoch ' + str(epoch)))
plt.plot(X.numpy(), Y.numpy(), 'r')
if leg == True:
plt.legend()
else:
passSet the random seed:
# Set random seed
torch.manual_seed(0)<torch._C.Generator at 0x7c89b4a521f0>Get Some Data
Create the Data class
# Create the data class
class Data(Dataset):
# Constructor
def __init__(self):
self.x = torch.arange(-1, 1, 0.1).view(-1, 1)
self.y = torch.zeros(self.x.shape[0], 1)
self.y[self.x[:, 0] > 0.2] = 1
self.len = self.x.shape[0]
# Getter
def __getitem__(self, index):
return self.x[index], self.y[index]
# Get Length
def __len__(self):
return self.lenMake Data object
# Create Data object
data_set = Data()Create the Model and Total Loss Function (Cost)
Create a custom module for logistic regression:
# Create logistic_regression class
class logistic_regression(nn.Module):
# Constructor
def __init__(self, n_inputs):
super(logistic_regression, self).__init__()
self.linear = nn.Linear(n_inputs, 1)
# Prediction
def forward(self, x):
yhat = torch.sigmoid(self.linear(x))
return yhatCreate a logistic regression object or model:
# Create the logistic_regression result
model = logistic_regression(1)Replace the random initialized variable values with some predetermined values that will not converge:
# Set the weight and bias
model.state_dict() ['linear.weight'].data[0] = torch.tensor([[-5]])
model.state_dict() ['linear.bias'].data[0] = torch.tensor([[-10]])
print("The parameters: ", model.state_dict())The parameters: OrderedDict({'linear.weight': tensor([[-5.]]), 'linear.bias': tensor([-10.])})Create a plot_error_surfaces object to visualize the data space and the parameter space during training:
# Create the plot_error_surfaces object
get_surface = plot_error_surfaces(15, 13, data_set[:][0], data_set[:][1], 30)<Figure size 640x480 with 0 Axes>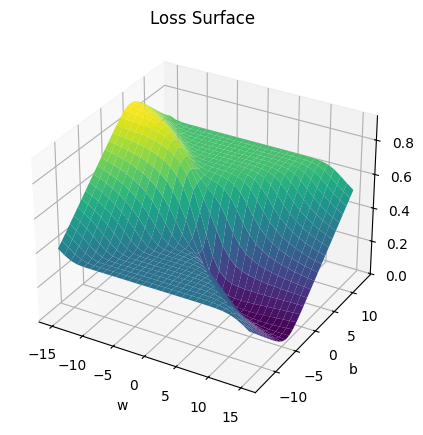
Define the dataloader, the cost or criterion function, the optimizer:
# Create dataloader object, crierion function and optimizer.
trainloader = DataLoader(dataset=data_set, batch_size=3)
criterion_rms = nn.MSELoss()
learning_rate = 2
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)Train the Model via Batch Gradient Descent
Train the model
# Train the model
def train_model(epochs):
for epoch in range(epochs):
for x, y in trainloader:
yhat = model(x)
loss = criterion_rms(yhat, y)
optimizer.zero_grad()
loss.backward()
optimizer.step()
get_surface.set_para_loss(model, loss.tolist())
if epoch % 20 == 0:
get_surface.plot_ps()
train_model(100)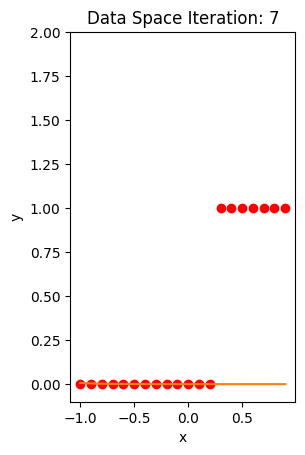
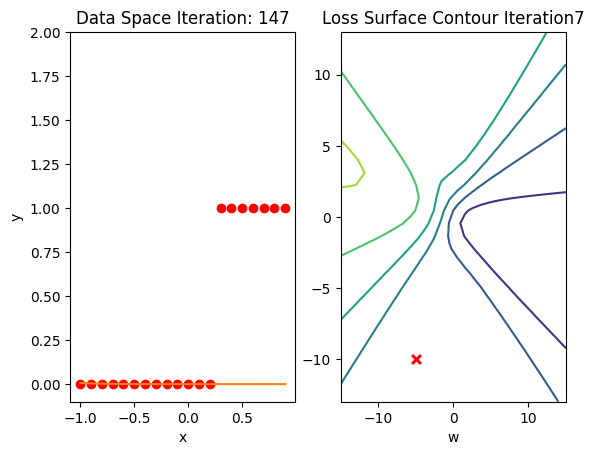
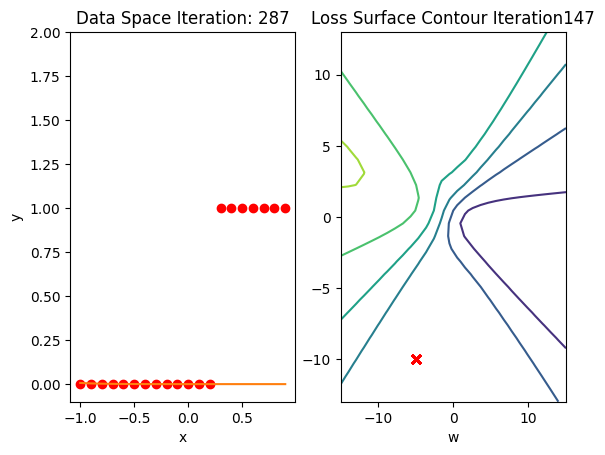
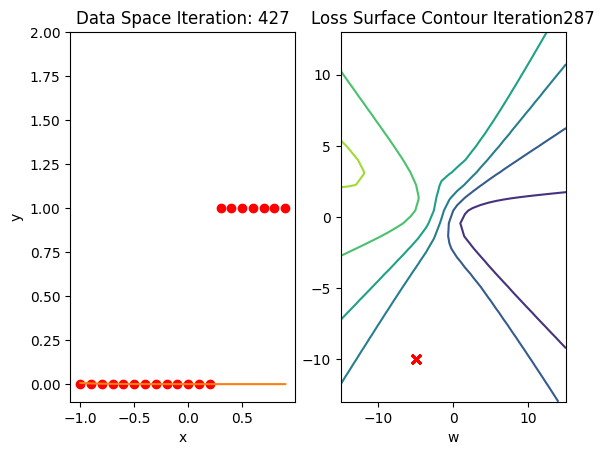
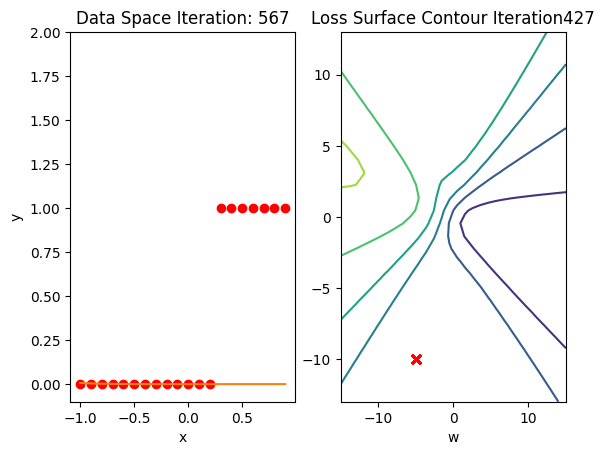
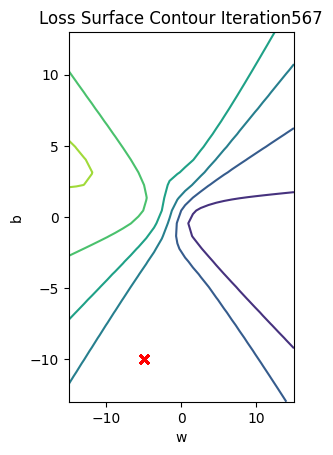
Get the actual class of each sample and calculate the accuracy on the test data:
# Make the Prediction
yhat = model(data_set.x)
label = yhat > 0.5
print("The accuracy: ", torch.mean((label == data_set.y.type(torch.ByteTensor)).type(torch.float)))The accuracy: tensor(0.6500)Accuracy is 60% compared to 100% in the last lab using a good Initialization value.